Aashish Jain and Professor Kihara have recently developed Phylo-PFP
09-14-2018
Function annotation of proteins is fundamental in biology. Predicting the function of genes/proteins can provide clues for interpreting experimental data as well as assist biologists to build hypotheses for designing experiments.
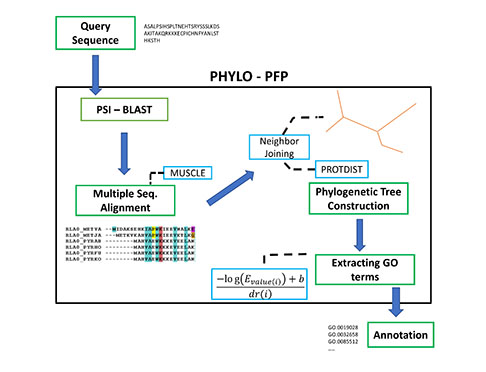
Aashish Jain and Professor Kihara have recently developed Phylo-PFP, a new sequence-based protein function prediction method, which mines functional information from a broad range of sequences including sequences with weak similarity with the query sequence. They first confirmed that the sequence similarity criteria (E-value) used by BLAST, a conventional bioinformatics tool for function annotation, does not often agree with the evolutionary distance defined by a phylogenetic tree. This discrepancy is one of the sources of errors in function annotation.
Thus, the new method, Phylo-PFP, combines a phylogenetic tree a PSI-BLAST search, which is shown to yield higher function annotation accuracy than existing methods.
Phylo-PFP is provided as an online server for public use: http://kiharalab.org/phylo_pfp.php
This work was published on August 25, 2018, in Oxford Bioinformatics:
https://doi.org/10.1093/bioinformatics/bty704
Contact: Daisuke Kihara http://kiharalab.org
Funding source: The Office of the Director of National Intelligence (ODNI), Intelligence Advanced Research Projects Activity (IARPA).