Structural Modeling of Protein Complexes with Disordered Proteins
06-16-2017
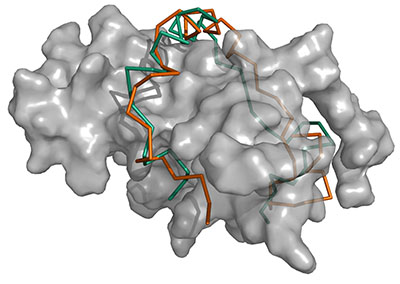
A substantial fraction of the proteins encoded in genomes are intrinsically disordered proteins (IDPs), which lack a single stable structure in the native state. IDPs serve many functions including mediating protein-protein interactions (PPIs). Such disordered PPIs are prevalent in important regulatory pathways, including many interactions of the tumor suppressor protein p53. To elucidate the molecular mechanisms of disordered PPIs, obtaining tertiary structure information is essential; however, they are difficult to study with experimental techniques and existing computational protein-protein and protein-peptide modeling methods are unable to model disordered PPIs. Here we present a novel computational method for modeling the structure of disordered PPIs, which is the first of this sort. The method, IDP-LZerD, is designed to follow a known biophysical picture of the mechanism of how IDPs interact with structured proteins. IDP-LZerD successfully modeled the majority of disordered PPIs tested. This technique opens up new possibilities for structural studies of IDPs and their interactions.
The paper was published on PLOS Computational Biology.
Paper: https://doi.org/10.1371/journal.pcbi.1005485
Modeling disordered protein interactions from biophysical principles. Lenna X. Peterson, Amitava Roy, Charles Christoffer, Genki Terashi, Daisuke Kihara, 2017, 13(4):e1005485. doi: 10.1371/journal.pcbi.1005485.
Contact: Daisuke Kihara http://kiharalab.org