Leung lab established a statistical approach to analyze zebrafish locomotor behaviour
06-19-2017
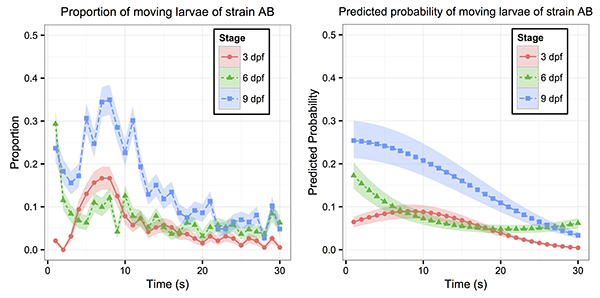
Our lab focuses on finding new drugs for retinal degeneration through zebrafish research. When we evaluate visual performance of zebrafish larvae after drug treatment, we would expose them to drastic light change. This light change would stimulate the larvae with normal vision to swim more than those with retinal degeneration. We can simultaneously measure this swimming (locomotor) behaviour from many larvae positioned in multi-well plates, which enables us to screen many drugs. However, this data collection scheme poses several statistical challenges. For example, some larvae display little or no movement during the short data-collection interval. Thus, the larval responses have many zero values and are imbalanced. These responses are also measured repeatedly from the same well during experiments, which results in correlated observations. To address these statistical issues, we worked with Yiwen Liu and Dr. Ping Ma from the Department of Statistics at the University of Georgia, and established a generalized linear mixed model (GLMM). The GLMM can effectively handle the data-imbalance and location-correlation issues. Using this GLMM approach, we can quantify true biological effects on zebrafish locomotor response, and can efficiently use this behaviour to find new drugs for retinal degeneration.
Reference:
Liu Y, Ma P, Cassidy PA, Carmer R, Zhang G, Venkatraman P, Brown SA, Pang CP, Zhong W, Zhang M, Leung YF. Statistical Analysis of Zebrafish Locomotor Behaviour by Generalized Linear Mixed Models. Sci Rep. 2017 Jun 7;7(1):2937.