New mechanism to destroy viruses could lead to future therapies
07-20-2017
Writer(s): Kayla Zacharias
WEST LAFAYETTE, Ind. — Researchers at Purdue University have uncovered the way some antibodies neutralize infections caused by viruses.
Bacterial and viral infections are similar in many ways and often cause comparable symptoms in humans. However, bacterial infections can frequently be treated with antibiotics, whereas viral infections are often not so easily cured.
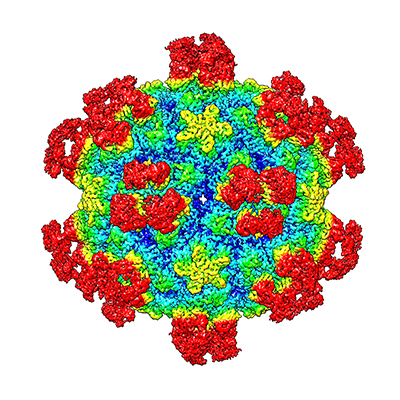
The research was published in Proceedings of the National Academy of Sciences and authored by Yangchao Dong, Yue Liu, Wen Jiang, Thomas J. Smith, Zhikai Xu and Michael G. Rossmann. The study shows that certain antibodies have the ability to open up a virus and empty its genome. This would be the equivalent of killing it, if viruses were alive in the first place.
“We’re anxious to understand how common cold viruses infect a host,” said Michael Rossmann, lead researcher and Hanley Distinguished Professor of Biological Sciences. “This gives us some idea of how the genome gets released once the virus has recognized the host cell.”
The study shows that binding of a neutralizing antibody to a specific rhinovirus causes significant changes around holes in the shell of the virus, allowing the viral RNA to escape. This “uncoating” process halts the virus’ ability to replicate itself in the cell.
“Normally in the infectious cycle, the virus has to release its genome into the cell so that it can replicate,” said Liu, a postdoctoral researcher at Purdue. “On the surface of cells, there are a number of receptor molecules that can trigger the release of the virus genome. Now we’ve found an antibody that can do the same thing.”
The team used cryo-electron microscopy (cryo-EM), an imaging method used to create three-dimensional images of microscopic objects, to illustrate the process. These structures are the highest resolution image of an antibody-virus complex to date, according to the study.
There has been a “resolution revolution” in the last three years or so, said Rossmann. The invention of new detectors used in electron microscopes has enabled researchers to use cryo-EM when they would have used x-ray crystallography in the past. While it might take one or two years to capture a three-dimensional image using x-ray crystallography, it would take a few weeks using cryo-EM.
“From this, we know a lot of atomic detail about how the antibody interacts with the virus in terms of amino acids, chemical interactions and how the virus has changed its structure and prepared itself for genome release,” said Liu. “Without a high-resolution structure, we wouldn’t know all of these details.”
Although the results of the study don’t provide a direct cure for the common cold, they do indicate that scientists are getting closer. Now that they know how an antibody can neutralize viral infection, they can apply the mechanism to similar viruses.
“We now have two examples of antibody-induced genome release from the capsid, so we can utilize this mechanism to design the basic structure of the antigen,” Liu said. “These could be used to simulate the production of neutralizing antibodies that might be useful in controlling similar viruses.”
The study was funded by grant AI011219 from the National Institutes of Health. Dong, a visiting scholar from China, was supported by the China Scholarship Council.
Writer: Kayla Zacharias, 765-494-9318, kzachar@purdue.edu
Sources: Michael Rossmann, 765-494-4911, mr@purdue.edu
Yue Liu, yue.liu.6@purdue.edu
ABSTRACT - Antibody-induced uncoating of human rhinovirus B14
Yangchao Dong1, Yue Liu1, Wen Jiang1, Thomas J. Smith2, Zhikai Xu3, and Michael G. Rossmann1
1Purdue University, West Lafayette, IN; 2Fourth Military Medical University, Xi’an, China; 3University of Texas Medican Branch at Galveston, TX
Rhinoviruses (RVs) are the major causes of common colds in humans. They have a nonenveloped icosahedral capsid surrounding a positive-strand RNA genome. Here we report that the antigen-binding (Fab) fragment of a neutralizing antibody (C5) can trigger genome release from RV-B14 to form emptied particles and neutralize virus infection. Using cryo-electron microscopy, structures of the C5 Fab in complex with the full and emptied particles have been determined at 2.3 Å and 3.0 Å resolution, respectively. Each of the 60 Fab molecules binds primarily to a region on viral protein 3 (VP3). Binding of the C5 Fabs to RV-B14 results in significant conformational changes around holes in the capsid through which the viral RNA might exit. These results are so far the highest resolution view of an antibody–virus complex and elucidate a mechanism whereby antibodies neutralize RVs and related viruses by inducing virus uncoating.