Links between DNA methylation and nucleosome occupancy in the human genome Clayton K. Collings and John N. Anderson
04-19-2017
Epigenetics & Chromatin 2017 10:18 DOI: 10.1186/s13072-017-0125-5
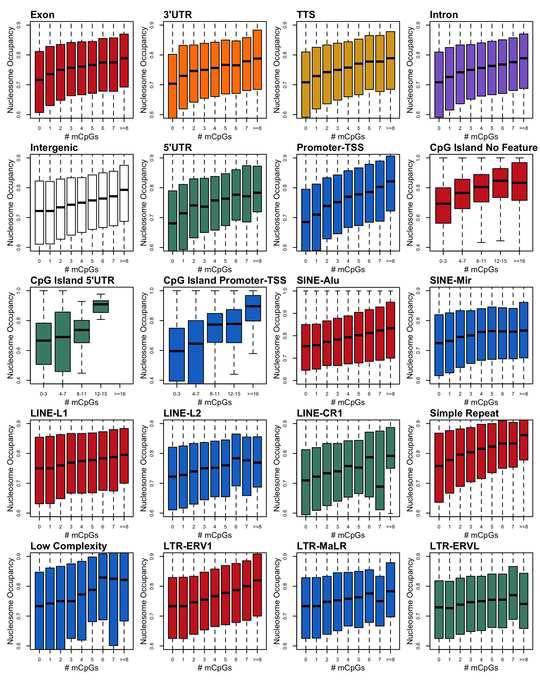
Previous studies from our laboratory have shown that DNA methylation directly enhances the stability of nucleosomes in vitro. However, the relationship between DNA methylation and nucleosome occupancy in the cell is poorly understood. In this study, we implemented a bioinformatics approach to study links between DNA methylation and nucleosome occupancy throughout the entire human genome. Using this approach, we demonstrated that increasing methylated CpG density is correlated with nucleosome occupancy and that in methylated CpG-rich nucleosomes, methylation levels are greater in the core than in the adjacent linker DNA. These nucleosomal DNA methylation patterns were detected not only in total genomic DNA but also within most subgenomic regions. Prominent exceptions to the positive correlation between mCpG density and nucleosome occupancy included CpG islands marked by H3K27me3 and CpG-poor heterochromatin marked by H3K9me3, and these modifications, along with DNA methylation, are the major silencing mechanisms of the human genome.