High-throughput discovery of ligand binding proteins in a proteome
02-13-2017
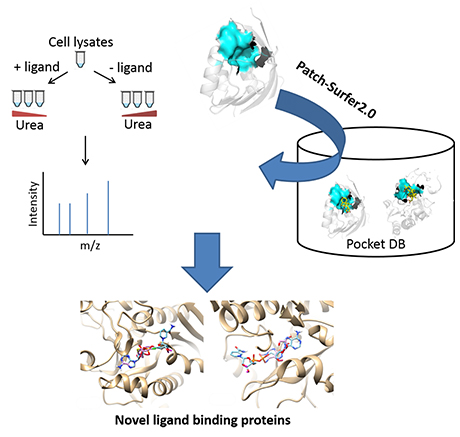
Detecting interaction networks of molecules is an important step for systematic understanding of molecular mechanisms of living cells. While protein-protein interaction networks have been identified in many organisms, methodology for revealing protein-ligand interaction networks is not well established yet.
In this work, researchers in the Kihara lab, Drs. Woong-Hee Shin, Xiaolei Zhu, and the PI, Dr. Kihara, in collaboration with the Tao Lab (Biochemistry) and the Chiwook Park lab (MCMP), developed an integrated experimental and computational approach to systematically identify proteins that bind a ligand molecule in a cell. The approach was applied to find novel nicotinamide adenine dinucleotide (NAD)-binding proteins in the Escherichia coli proteome.
For the experimental part, pulse proteolysis was coupled with filter-assisted sample preparation (FASP) and quantitative mass spectrometry. Under denaturing conditions, ligand binding affected protein stability, which resulted in altered protein abundance after pulse proteolysis. For the computational part, we used the software Patch-Surfer2.0 developed in the Kihara group, which predicts binding ligands for a protein by comparing the shape and physicochemical properties of potential binding sites of the protein to known ligand binding pockets in a database. By taking consensus of proteins detected by the experimental and the computational methods 8 proteins were identified that are not previously known as NAD binders. Further validation of these 8 proteins showed that their binding affinities to NAD are higher than their cognate ligands. The 8 proteins also have consistent experimental profile to known NAD-binders. These results strongly suggest that these eight proteins are indeed newly identified NAD binders.
The method has a wide range of applications including identifying drug binding proteins in human cells.
This work was reported on the current (February 2017) issue of Journal Proteome Research.
Paper:http://pubs.acs.org/doi/abs/10.1021/acs.jproteome.6b00624
Discovery of Nicotinamide Adenine Dinucleotide Binding Proteins in the Escherichia coli Proteome Using a Combined Energetic- and Structural-Bioinformatics-Based Approach. Zeng L, Shin WH, Zhu X, Park SH, Park C, Tao WA, Kihara D. J Proteome Res. 2017 Feb 3;16(2):470-480.
Source: Daisuke Kihara http://kiharalab.org