Delving into Host-Parasite Interactions
08-16-2016
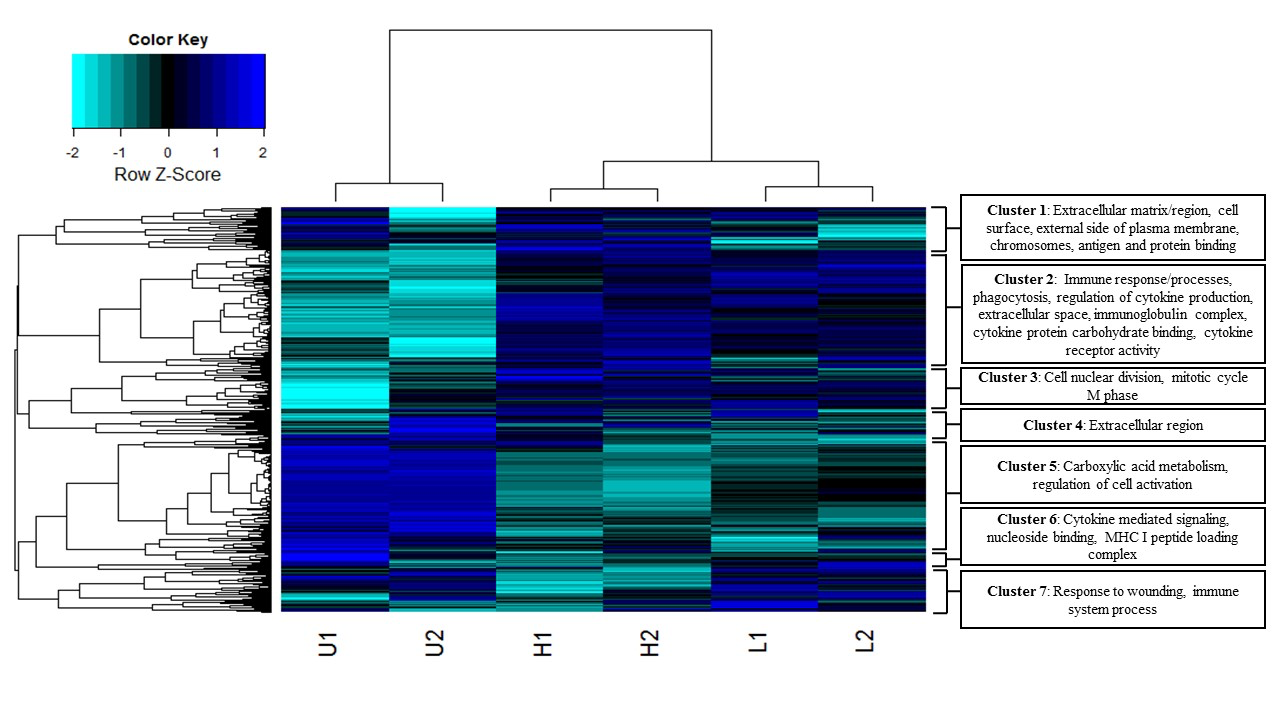
In a recent paper in BMC Genomics (2016) 17:600, former EEB graduate student Bhagya Wijayawardena, and her co-advisors D.J. Minchella and J.A. DeWoody explore the influence of trematode parasite burden on gene expression in a mammalian host.
Parasites can profoundly impact their hosts and are responsible for a plethora of debilitating diseases. To identify global changes in host gene expression related to parasite infection, we sequenced, assembled, and annotated the liver transcriptomes of Balb/cj mice infected with the trematode parasite Schistosoma mansoni and compared the results to uninfected mice. We used two different methodologies (i.e. de novo and reference guided) to evaluate the influence of parasite sequences on host transcriptome assembly. Overall, our data indicate that a) infected mice reduce the expression of key metabolic genes in direct proportion to their infection level; b) infected mice similarly increase the expression of key immune genes in response to infection; c) patterns of gene expression correspond to the pathological symptoms of schistosomiasis; and d) identifying and filtering out non-target sequences (xenobiotics) improves differential expression prediction. Our findings identify parasite targets for RNAi or other therapies and provide a better understanding of the pathology and host immune repertoire involved in response to S. mansoni infections.