Thinking ahead: how a unicellular nitrogen fixing cyanobacterium manages its gene expression to accommodate the challenges it faces through out a 24-hour day
02-12-2015
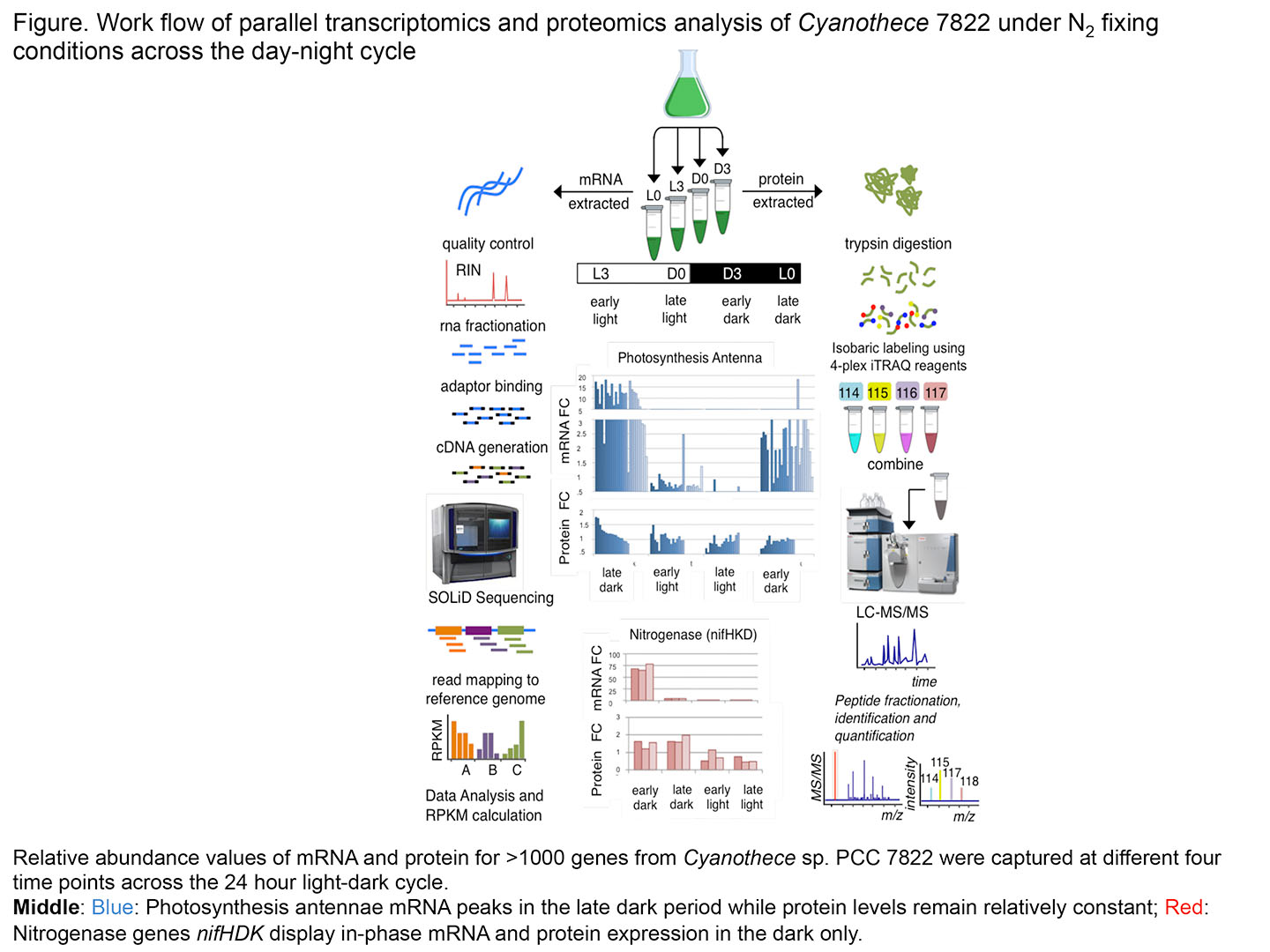
Cyanobacteria are important model organisms that use sunlight to power photosynthesis and some even fix N2 from the atmosphere to make proteins and DNA. In addition, cyanobacteria are the only prokaryotes known to have a circadian clock that helps regulate gene expression throughout the day-night cycle. One of our objectives is to understand how the circadian clock helps determine when genes are turned on and off and the impact that this mechanism has on metabolites that might be useful as biofuels.
We combined mass-spectrometry based proteomics and RNA-sequencing transcriptomics (RNA-Seq) to quantitatively measure a total of 6766 mRNAs and 1748 proteins at four time points across a 24 hour light–dark cycle (procedure diagramed in the figure). We were able to learn how the expression of the main metabolic pathways (glycolysis, photosynthesis, TCA, nitrogen fixation, etc.), as well as specific pathways relevant to biofuels, fluctuate during the light–dark cycle.
One of the most interesting highlights of this work was that the cell could anticipate future metabolic needs in the next daily phase. Thus, mRNA levels rose in the late dark for proteins needed in the light. The expression of the photosynthesis antennae is one example of this phenomenon. Most of the genes are transcribed in the dark and the proteins are fairly stable throughout the 24-hour period. A second key finding was that protein levels do not fluctuate nearly as much as do mRNA levels. Specific proteins such as those involved in N2 fixation (night) and photosynthesis (day) are present only at specific times during the cycle. However some 90% of the proteins vary little in amount throughout the day-night cycle as the cell conserves energy.
Revealing all the complex ways different metabolic pathways interact and play off of each other in these microorganisms is important for future research. The results will permit us to design better strategies for engineering this strain to perform useful tasks, such as producing biofuels or other beneficial chemicals. We also learned more about how these microbes regulate their energy levels and deal with the challenging balancing act of efficiently performing photosynthesis and nitrogen fixation in the same cell. This work was funded in part by a grant from the Office of Science (BER), U. S. Department of Energy.
Welkie D, Zhang X, Markillie ML, Taylor R, Orr G, Jacobs J, Bhide K, Thimmapuram J, Gritsenko M, Mitchell H, Smith RD, Sherman LA: Transcriptomic and proteomic dynamics in the metabolism of a diazotrophic cyanobacterium, Cyanothece sp. PCC 7822 during a diurnal light-dark cycle. BMC genomics 2014, 15:1185.
Article and figure submitted by Louis Sherman, Professor of Biological Sciences.